Drawing ecological inferences from coincident patterns of population- and community-level biodiversity
June 9, 2014 Leave a comment
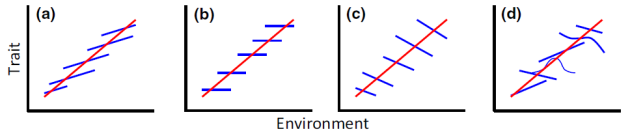
What’s your favourite intraspecific (blue) and interspecific/community-level (red) trait-environment relationship? Taken from Vellend et al.
I picked this paper and broke our 6-month run of old-school classics (sorry!) because the topic looked interesting, and Mark Vellend seems to write so many good reviews I couldn’t help myself. Like Lynsey, I thought the paper was going to take a few different directions, but I enjoyed it nonetheless.
Figure 1 (which I’ve put at the top of this article) is the sort of graph that lots of people seem to grappling with, and I’m glad to see it in print. It’s an extension of one from another excellent paper, and the essential point is that an inter-specific relationship could be the opposite of the intra-specific relationship among the same species. The similarities with Felsenstein’s classic graph showing why we need to control for phylogenetic history are striking – different results are found at different (phylo)genetic scales. If you view individuals as existing in a hierarchy from clade–>region(–>community?)–>species–>individual, then I think it’s amazing that we find these reversals at all scales. Why is it that ecological/evolutionary units seems to have discrete, real boundaries among them? I sense it’s related to the balance between character displacement (species spread out into discrete groups) and niche filling (within those groups there is variation), but I sense we need some more simulation analyses to truly understand why this keeps happening. I may try something over on my R blog next week (sorry for the plug!).
I found the sudden appearance of a meta-analysis later in the paper surprising but kind of cool. The general idea is that discrete fragments of habitat should show a greater potential for genetic drift than same-sized sections of continuous habitat, and changes in the correlation between genetic and species diversity reveal this. The results seem to support their conclusions, and (like all such provocative graphs) leave me wondering how further we could tease this all apart. Biogeographic history, generation time, age of habitat patch, etc., should all affect these sorts of correlations, and on second thought it somewhat shocks me that we don’t have better data on all this (“correlations were calculated from data not collected for this purpose“). If we are to stand a chance of understanding some of the hierarchical patterns that I described above, we need slightly more joined-up analyses of different levels of diversity. Indeed, there’s enough meta-data on GenBank that maybe someone (not me!) could have a go at this.
I was looking forward to this paper as I thought it would help clarify my own pet interest in integrating population and species level patterns of biodiversity and would give me more ammunition in my plot to have all above species level analyses incorporate intraspecific variation. The paper kind of managed to do this. It was slightly disconcerting how many times the authors stated what the paper was not going to talk about, but it did eventually end up focusing on congruent diversity patterns at the above (species numbers) and below (genetic diversity) species level and on congruent patterns of trait variation along environment gradients when considering multiple species or multiple populations of the same species. Both concepts sound fairly dull, let’s be honest, but have not been assessed vigourously in the literature so it was really nice to see a summary of these results. Conclusion: sometimes intra- and interspecies patterns are similar, sometimes not.
I guess where I felt the paper fell short (and this is probably due to me wanting the paper to be something it wasn’t trying to be) was that it did not really focus on how insights at one level of diversity could inform insights on another level. For instance, what are the characteristics (trait variation, genetic diversity) of populations that make up large range species vs. small range species? I.e. how do some species obtain large ranges, while others break up into multiple smaller ones? Are all large range species en route to becoming multiple small range ones or are there certain ‘types’ of population that can maintain a large range species over evolutionary time?
A ton of work has been done about edge and centre populations and adaption/movement in response to environmental change, I would have liked to have read more about how population characteristics can suggest how a species might respond (maladapted gene flow, good gene flow, rapid evolution, local extinction). Species’ ranges are dynamic because populations come and go. How do we integrate this knowledge with our desire to study emergent patterns above the species level (diversity fluxes at the regional scale, for instance)?
I did appreciate the authors’ discussion of trait vs. ‘diversity’ variation approaches to looking st intra/interspecific congruence. Molecular diversity measures are relatively easy to collect and more or less comparable across sampling units (a can of worms we won’t open here). Traits are harder to collect, it’s harder to choose relevant traits to collect in the first place and inference might be bamboozled if you are looking at a deficient trait or one collected poorly. I am guessing that a little way in the future, such studies will use the genetic variation behind the trait of interest (when genomes shower down on us like rain). Until then, a mix of both approaches is probably most informative.
Onwards and upwards.