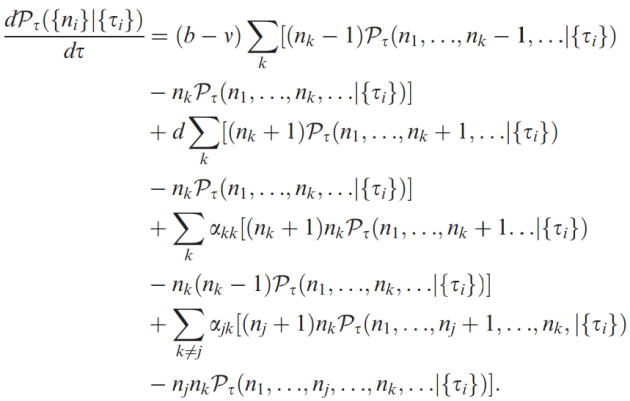Phylogenies support out-of-equilibrium models of biodiversity
April 13, 2015 Leave a comment
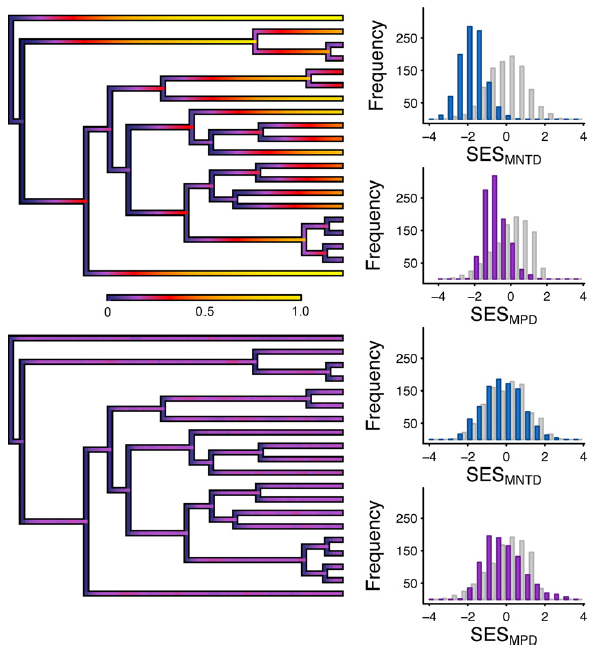
Overview of the new Speciation by Genetic Differentation model of Manceau et al.
It’s been a good few months for Neutral Theory in Ecology Letters (…and so, by extension, PEGE!). In this paper, Manceau et al. put forward an extension of Neutral Theory, including a new model of speciation (Speciation by Genetic Differentation), and relax the dependence upon a static metacommunity. Both are exciting extensions to the theory.
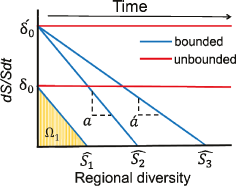
The concept of the meta-community is something that’s always troubled me about Neutral Theory, because it seems a bit much to appeal to something outside the system to keep the system stable. Yet at the same time the only thing I can think of that’s less realistic than a meta-community is not having a meta-community, since clearly no community evolves in isolation. In this model the meta-community can change through time (i.e., it’s not at equilibrium); it’s no longer a deus ex machina, and is instead a part of the ecological theatre (see what I did there? :p).
Equally, the speciation mechanism the authors put forward is a useful development. I’m a fan of protracted speciation models, but my general problem comes from fitting them onto a specific evolutionary process. I don’t doubt they describe pattern and process well, but they don’t seem to be linked to one process in particular. Thus the genetic differentation model the authors suggest is, to me, extremely exciting. As with all these exciting new models, it’s almost a shame that the cleverest bit – the maths – is too complex to present in the body of the paper (I say almost a shame, because I’m certain I wouldn’t understand it!).
To me, the most important concept in this paper is that telling phrase ‘out of equilibrium’. Arguments about whether diversification is or isn’t density-dependent are never going to go away, but there are some who are calling for the debate to happen in the context of recent ecological theory about what carrying capacities in systems look like. Personally, I think that a discussion on density-dependence has to happen with an understanding of species’ abundances, and that means individual-based models. Work like this is an important step towards this.
I must admit I read this paper far quicker than it merited, so my thoughts are a bit hazy and any qualms might be unfounded. On that note, here goes…
I did appreciate the positivity bouncing around in this paper. The authors were resolutely positive about the capacity of phylogenies and macroevolution in general to inform us on diversity patterns. This was nice to see as many people, myself included, often despair on the ability of phylogenies to tell us anything.
Their two tweaks to neutral theory also sound, on the whole, sensible tweaks that make sense given what we know about species and given that we agree we want to retain the simplicity of the neutral theory while identifying the key assumptions that make it fall down.
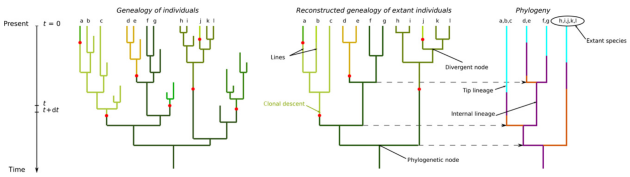
First, speciation by genetic differentiation. Indeed, closely related species generally do differ genetically. Whether this difference is just an accumulation of neutral mutations or some kind of adaptive divergence (and which of the two kinds is more common) is another question. I felt like the authors could have discussed this issue more deeply because as a naive reader I was left wondering about the biological reality of such an abstract speciation mechanism. Sure, the model does not claim to be 100% realistic, but a discussion of the different signatures expected depending on the speciation mode would have been nice. The authors talk a lot about future directions and models they would like to compare theirs too (e.g., Pigot’s biogeographic model) and I look forward to hearing about these extensions. They somewhat cryptically refer to some lineages acting like speciation hubs that presumably shoot out new species willy-nilly. What kind of lineages would these be? Large ranged? Sexually selected? Weird mating system? Host shifter?
Second, growing/shrinking metacommunity. I agree with the authors that a constant metacommunity seems unreasonable. But I would have liked to hear more about how a growing/shrinking metacommunity might come about. Are we talking about finer partitioning of a finite area or colonisation of new habitats or competitive exclusion of crappy species, or what? Is a metacommunity the right term to use when we are thinking about the interactions of ENTIRE species. Could populations of the same species occupy different metacommunities? (Meta-metacommunities!! :$).
My hunch is the authors have also thought of all of the above and this is just a first pass attempt, albeit an impressive one that (again) shows that with just a few small tweaks the overall premise of the neutral theory is really useful in understanding general diversity patterns. I remain on the fence whether all this tweaking is destroying the original premise of the neutral theory (as I see it, to provide a conceptually simple null with which we can work out which non-neutral processes really do matter).